Single Cell Multiomics
Single-cell RNA sequencing outshines conventional RNA sequencing in its capacity to identify cell-type specific changes in gene expression rather than average values in a bulk sample. Recent technological advancements in single-cell technologies have enabled the simultaneous measurement of different types of molecular data. Through multiomics analyses we can now gain transcriptomic, proteomic and epigenetic information from thousands of cells of different subsets in a single experiment.
Single-cell multiomics allows us to unravel the composition and function of various cell types within a highly complex sample, and thus to construct a more detailed map of the cellular and molecular signatures associated with pathophysiological processes or therapeutic intervention. Using this approach in the early stages of phase 0 studies can support the selection of the optimal biomarkers in your clinical study.
What we can add to your research
We help you to get a complete insight at the single-cell level.
CheckImmune utilizes leading droplet- and microwell-based technologies such as the 10x Genomics® Chromium™ and BD Rhapsody™ single-cell analysis systems, respectively, as well as efficient sequencing on the Illumina Novaseq platform.
We provide expert guidance on antibody panels, study design, and optimal sequencing parameters in order to get the most out of your precious samples.
We utilize robust and certified workflows using internal controls, targeted (rare) cell subset enrichment, sample multiplexing, and provide downstream data and statistical analysis tailored to satisfy your specific needs.
Would you like to talk to us?
Our Services
Consultancy for experimental design and planning
A modular single-cell sequencing toolbox to meet your research needs
- High-throughput droplet-based single-cell analysis
- Gentle microwell-based single-cell analysis
- Single-cell transcriptome
- Single-cell surface epitopes profiling
- Single-cell intracellular epitopes profiling
- Single-cell V(D)J immune repertoire analysis
- Single-cell open chromatin epigenetic profiling
- Cell hashing for sample multiplexing and cost reduction
Library preparation and sequencing
- Whole-transcriptome or targeted gene expression, epitopes, V(D)J, ATAC
- Illumina® sequencing
Processing and quality control of sequencing data
- Demultiplexing and read mapping using 10x Genomics Cell Ranger pipeline
- QC summary reports
- Raw sequencing data (FASTQ) and count matrix files
Downstream single-cell data analysis and interpretation (upon request)
- Using established R/Bioconductor workflows
- Custom analysis (visualization, clustering, annotation, DE, DA, GSEA, trajectory, ...)
- Support in scientific and medical interpretation
Equipment & Data Analysis:

10x Chromium Controller
BD Rhapsody
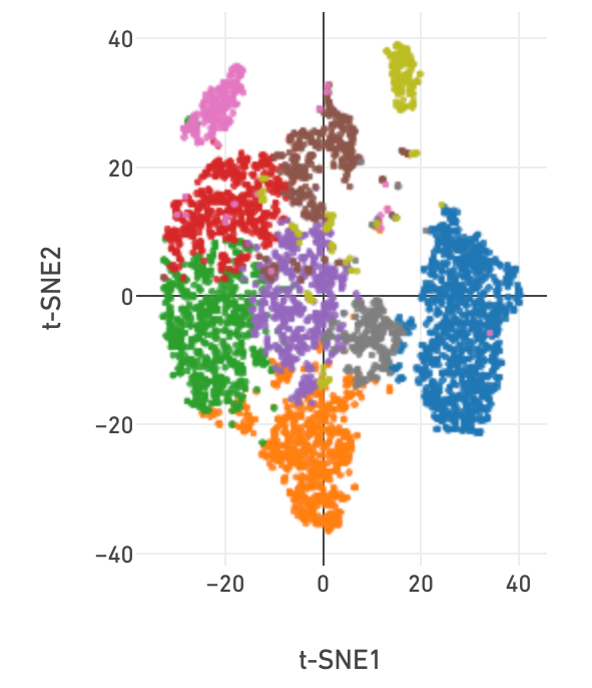
Single-Cell Genomics Pipelines

Established Workflows and Statistical Analysis
Key Information
- Single-cell transcriptome, surface epitopes, V(D)J profiling, chromatin accessibility
- High-quality multiomics single-cell sequencing workflows
- Expert support on single-cell experimental design, including strategies to analyse special on-target/off-target for (rare) cell subsets
- Cost-effective sequencing
- Demultiplexing and read mapping with standard 10x Genomics Cell Ranger or BD Single Cell Genomics pipelines
- Downstream single-cell data analysis (upon request)

